About ViBrism
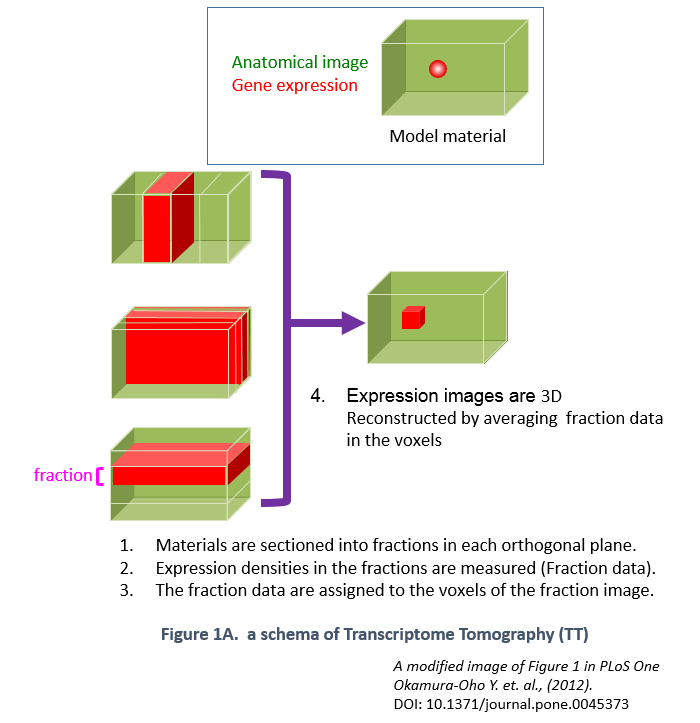
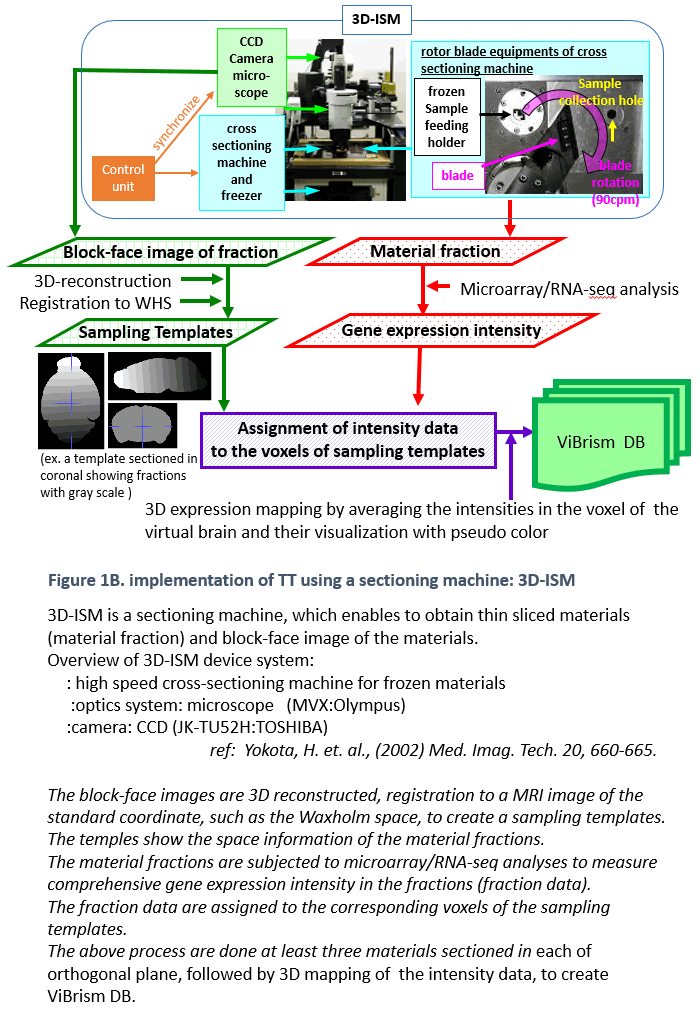
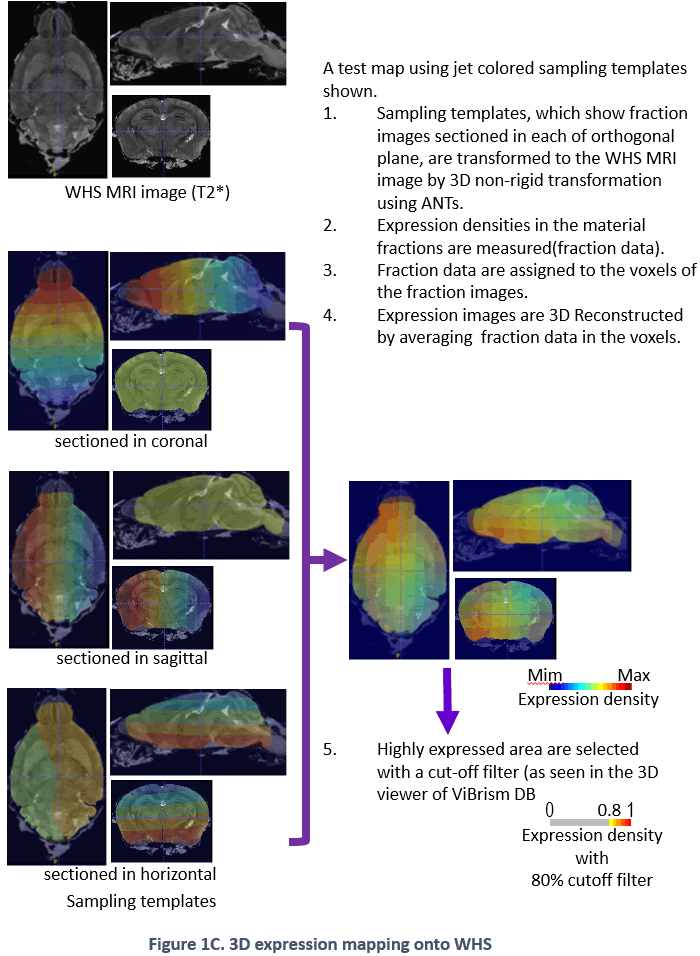
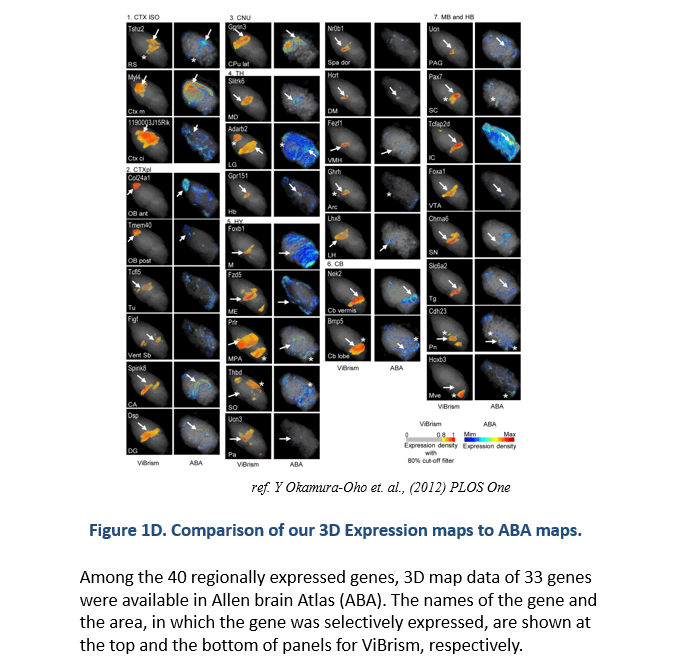
ViBrism (Virtual Brain with 3D-IDM) DB is a 3D-map database of endogenous gene expression in a virtual whole mouse brain produced with an original mapping framework Transcriptome Tomography (Figure 1: ref. PLoS One 2012, see a movie on rikenchannel-YouTube “Mapping genes onto brain structures”). 3D-reconstructed gene expression maps are displayed on the MRI image of the brain by image registration. ViBrism BD now contains 36,558 expression maps of the adult mouse brain (C57BL6/J 8-weeks male) and about 45,000 expression maps of each in the developmental stages (C57BL6/J 3 days, 7 days and 3weeks after birth, males), searchable by gene symbols (“Search by Gene symbol/ID”) or expression similarity (“Similarly Expressed Gene Search”) at our Top Page. After the search, 2D/3D expression maps and co-expression network graphs can be browsed and then data files are downloadable and viewed with a software on your desk-top computer (visit our “Software” page).